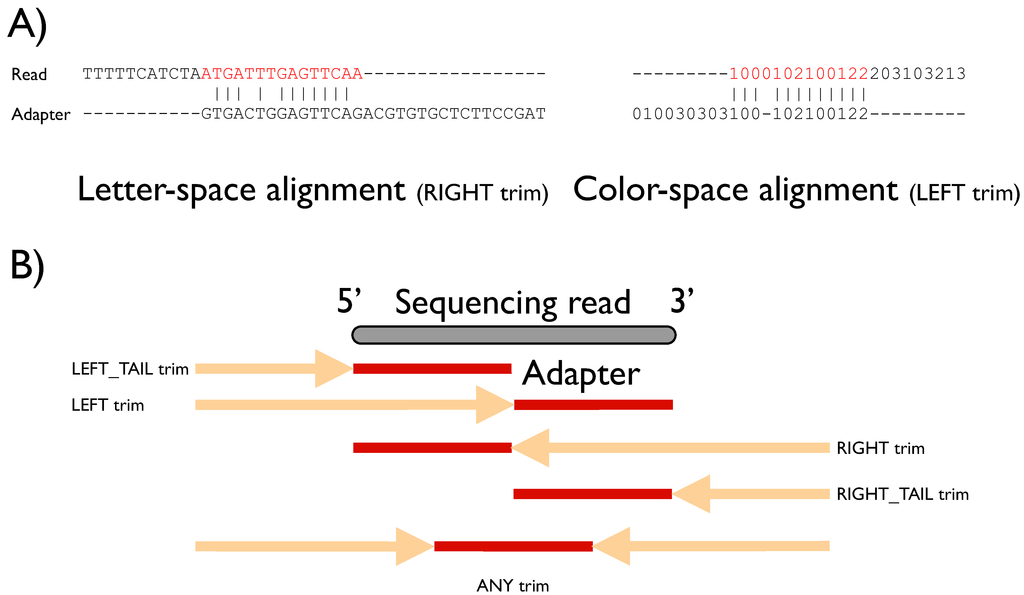
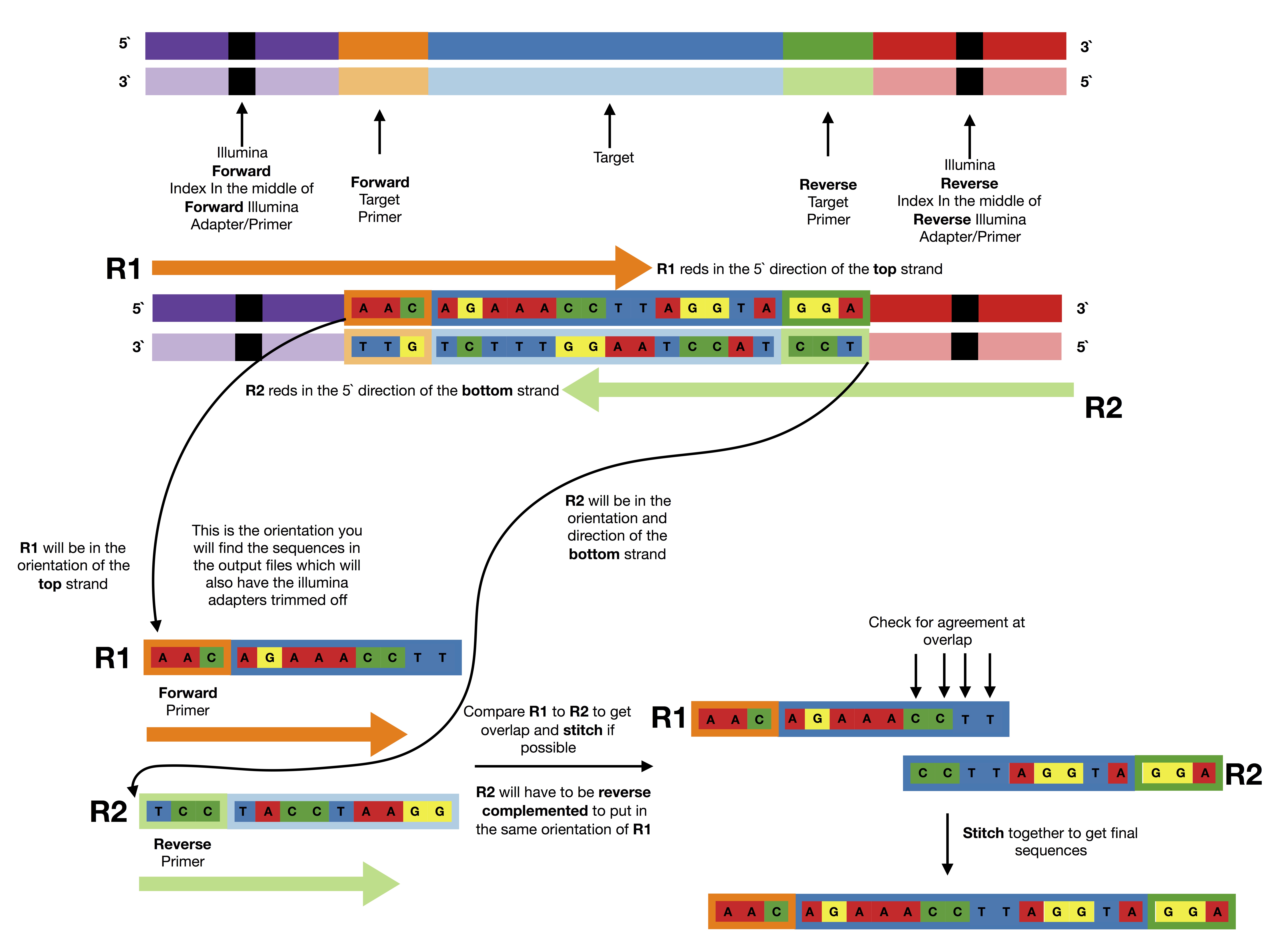
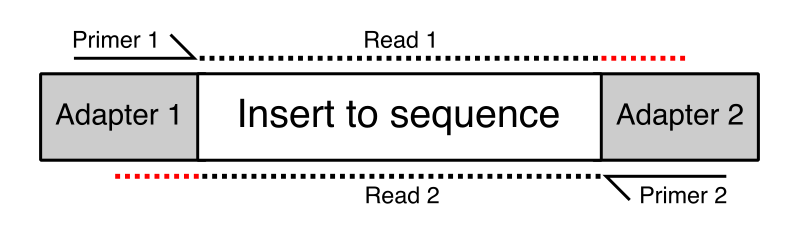
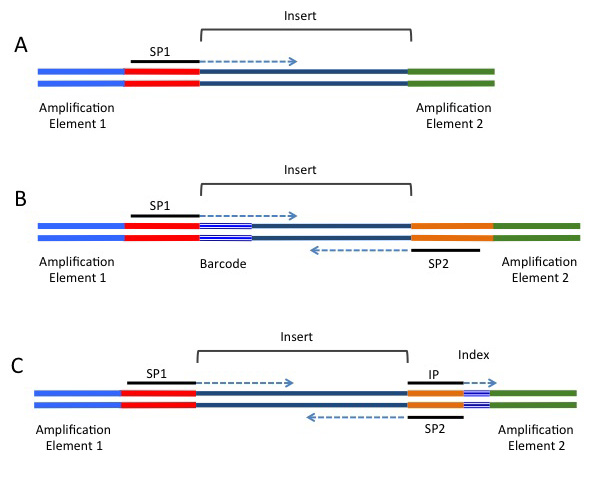
Adaptors and adaptor chimeras are a common sources of sequence artifacts | Download Scientific Diagram
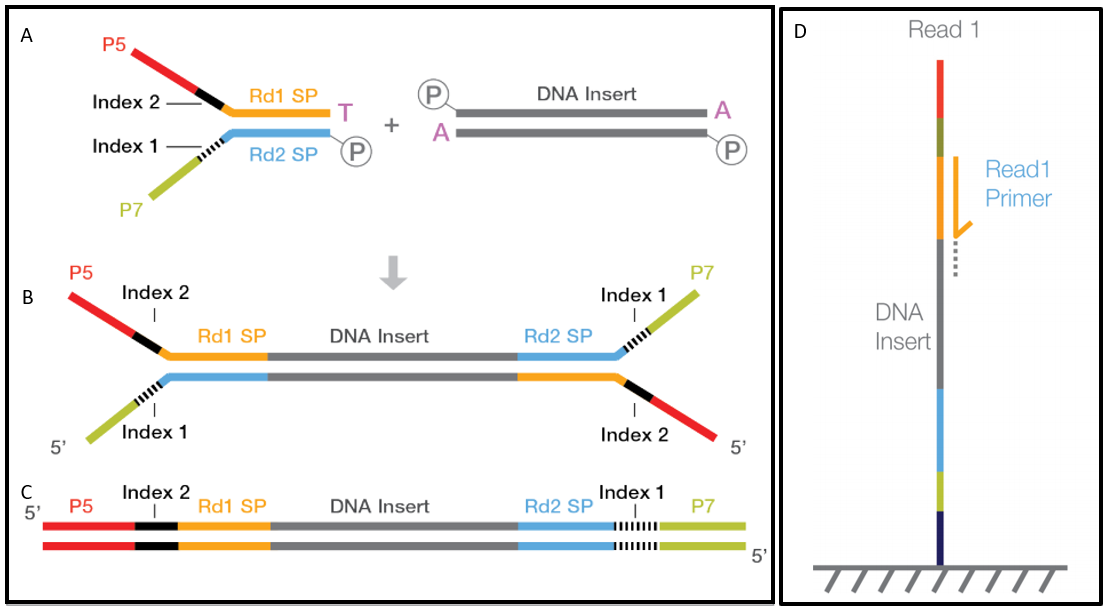
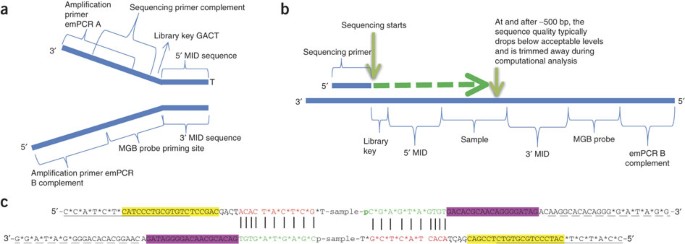
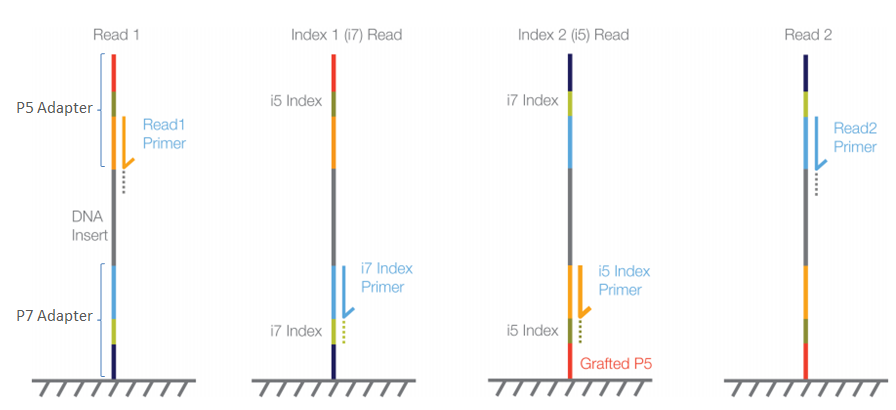
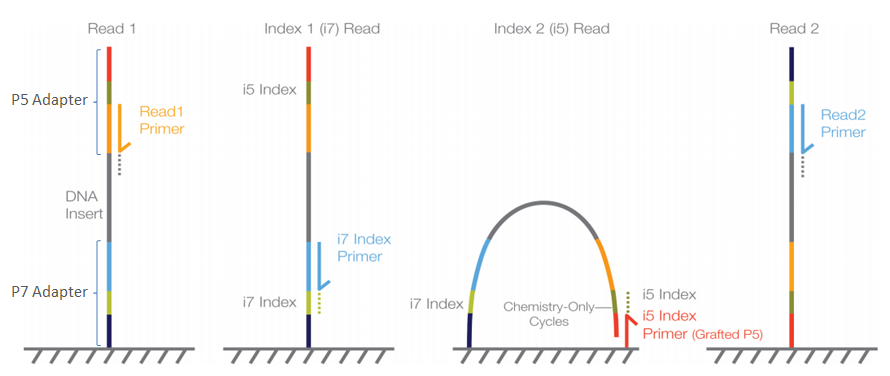
4-A scheme of targeted fragment being sequenced Sequencing adapters... | Download Scientific Diagram
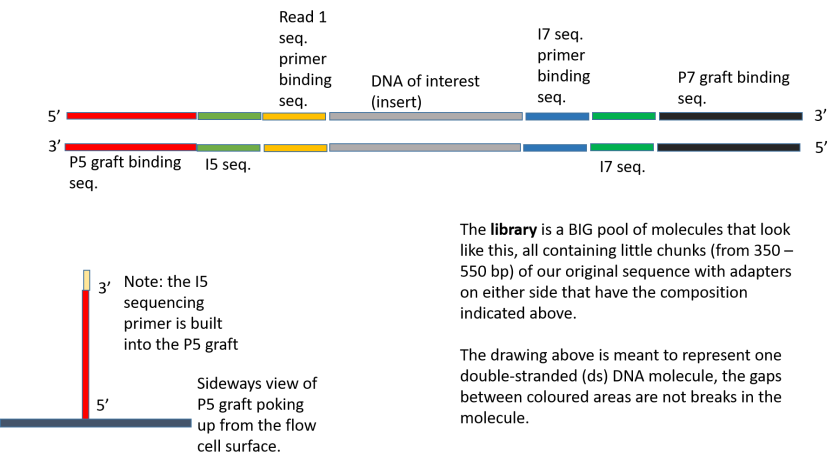
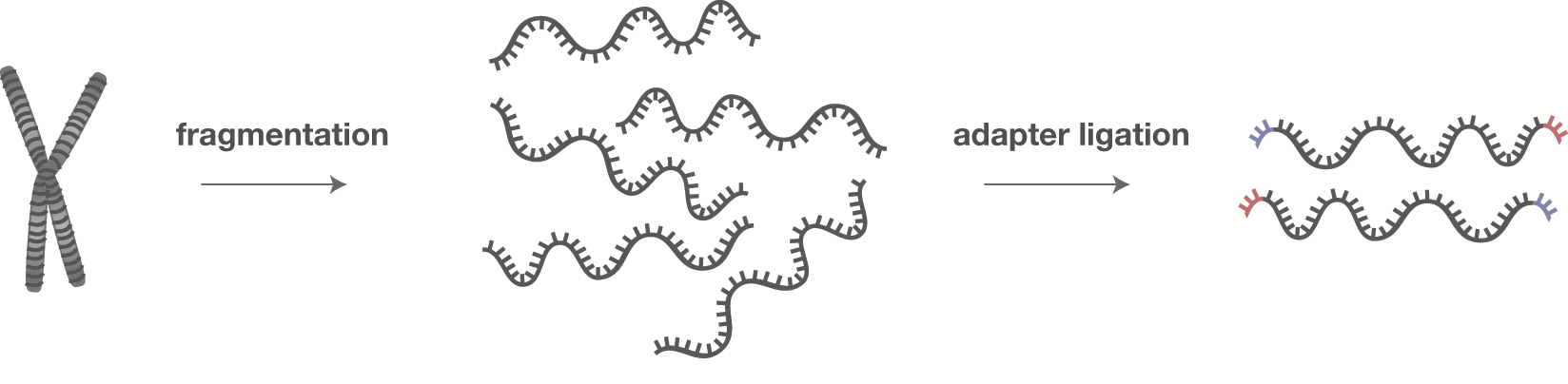
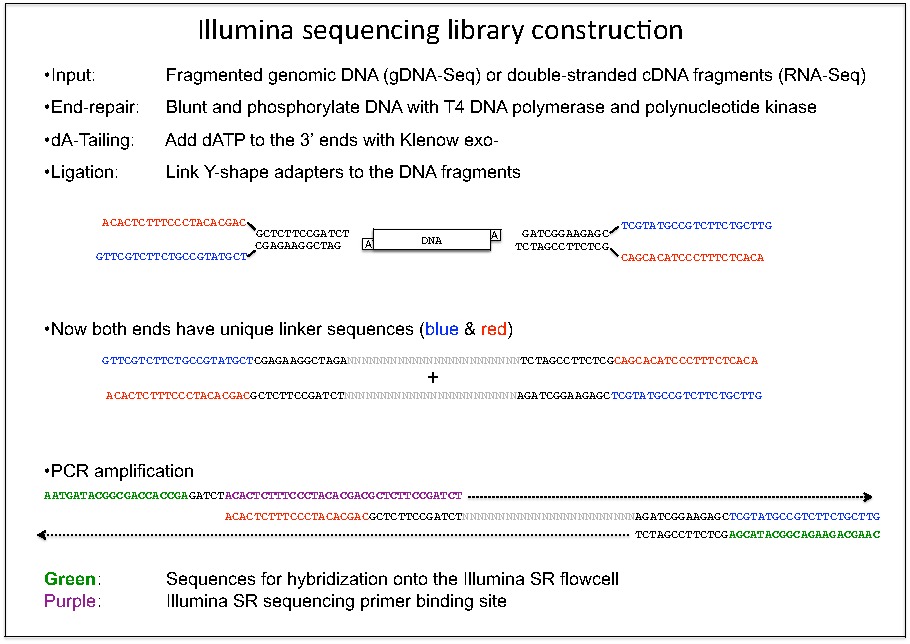
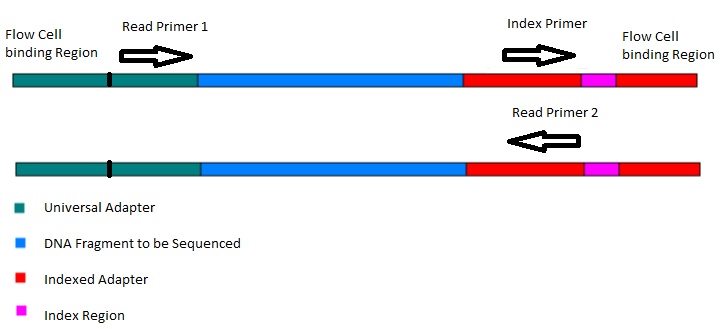
Illumina Sequencing (for Dummies) -An overview on how our samples are sequenced. – kscbioinformatics
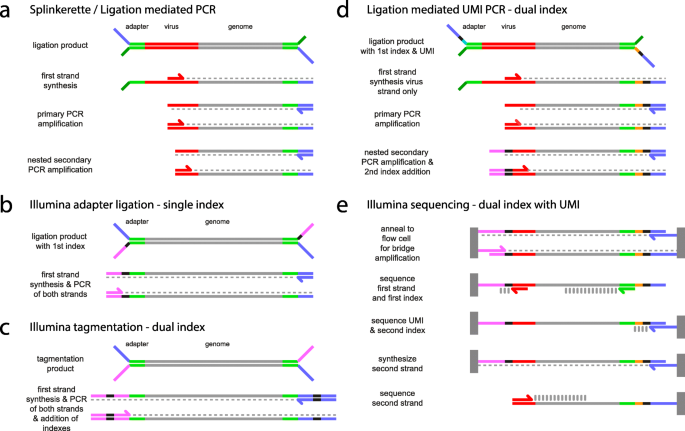
LUMI-PCR: an Illumina platform ligation-mediated PCR protocol for integration site cloning, provides molecular quantitation of integration sites | Mobile DNA | Full Text
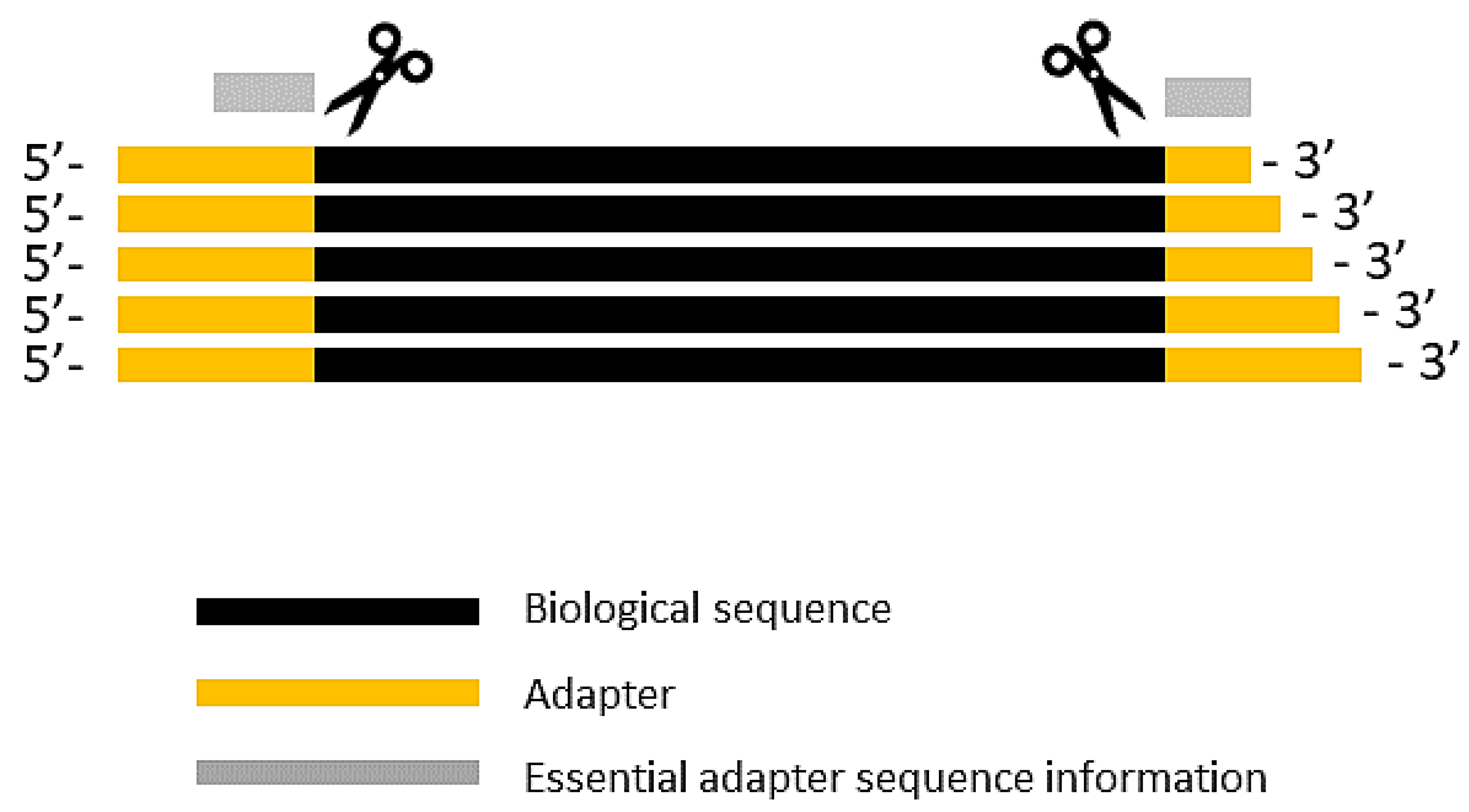
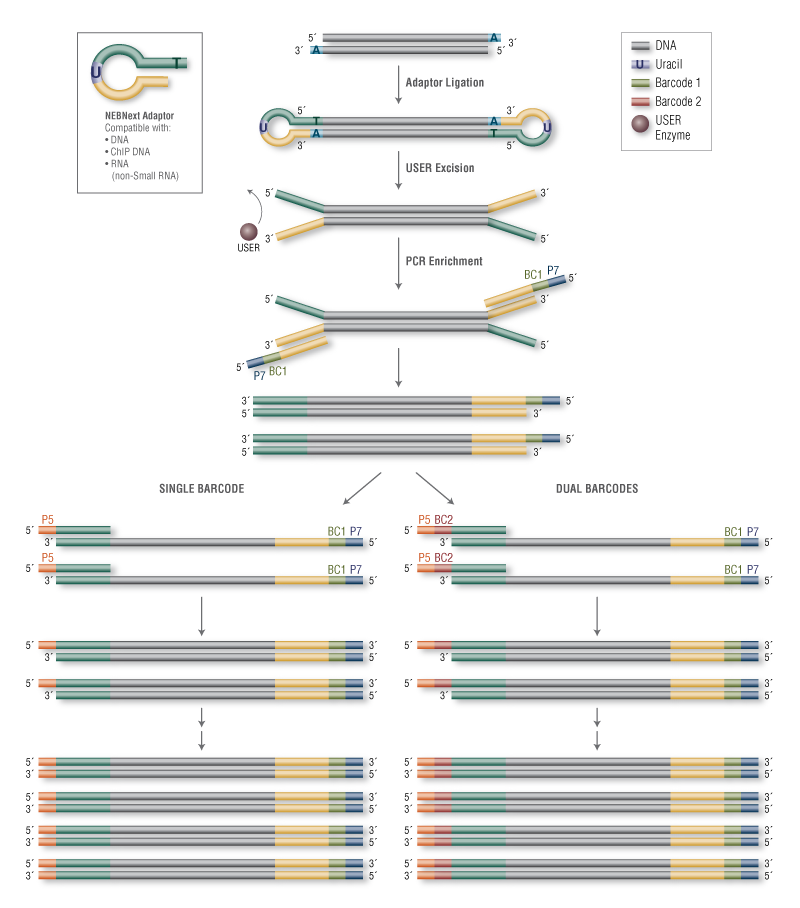
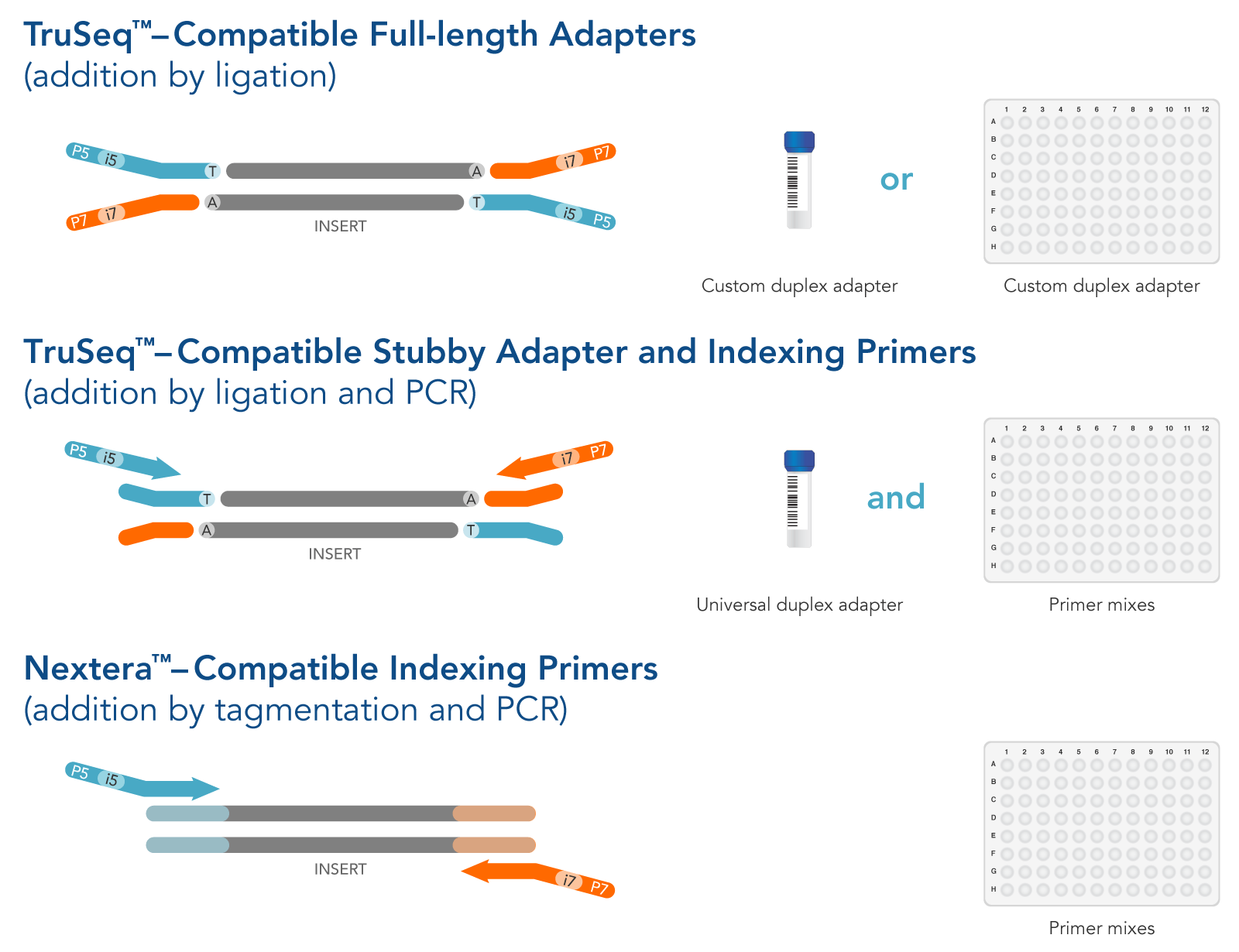
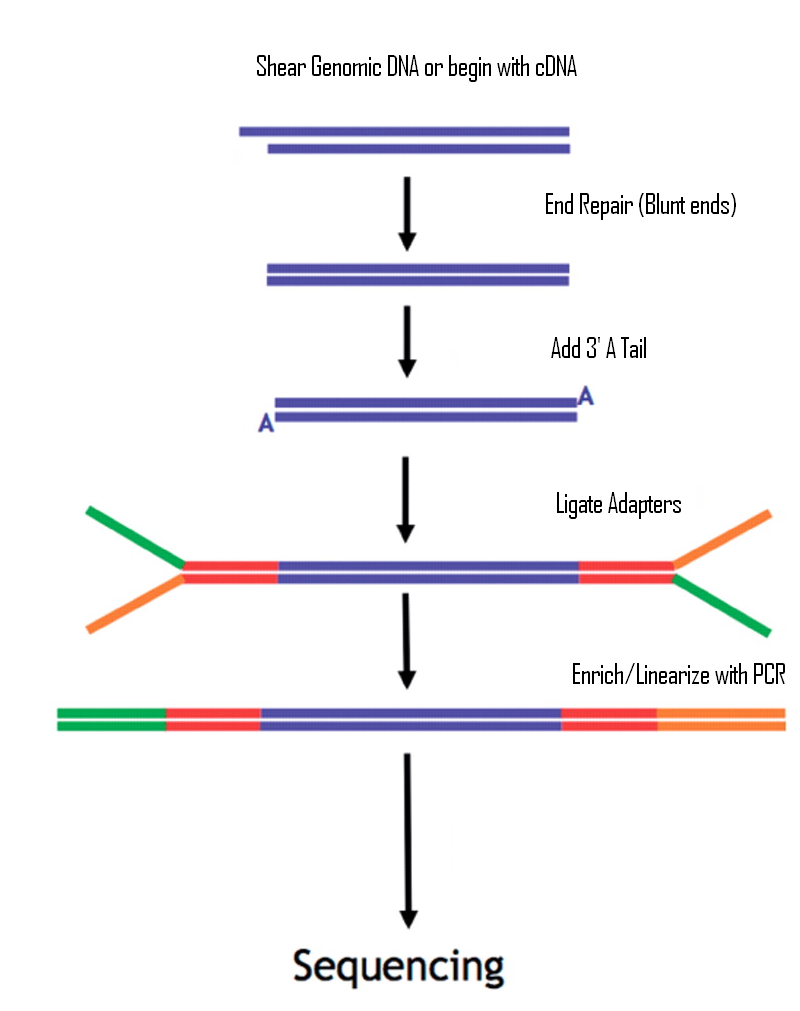
Biomolecules | Free Full-Text | High-Throughput Identification of Adapters in Single-Read Sequencing Data | HTML
High-Throughput, Amplicon-Based Sequencing of the CREBBP Gene as a Tool to Develop a Universal Platform-Independent Assay | PLOS ONE