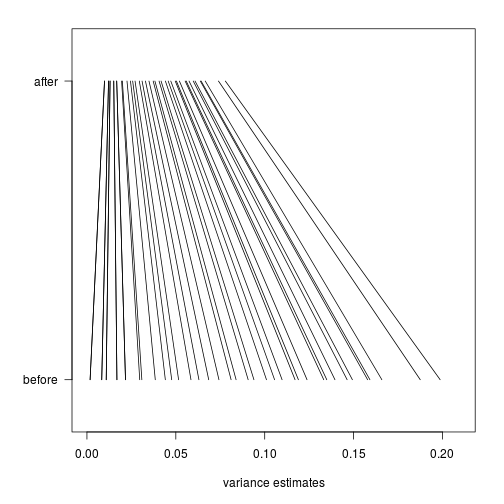
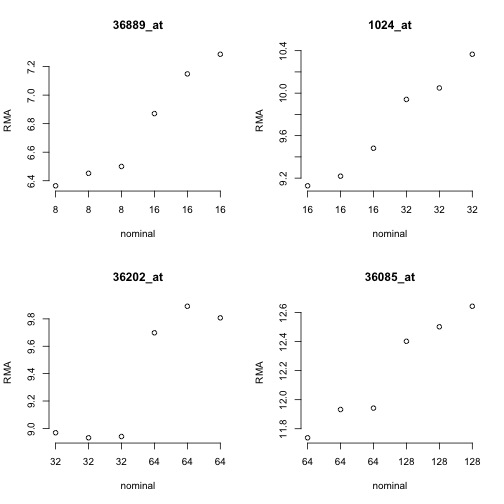
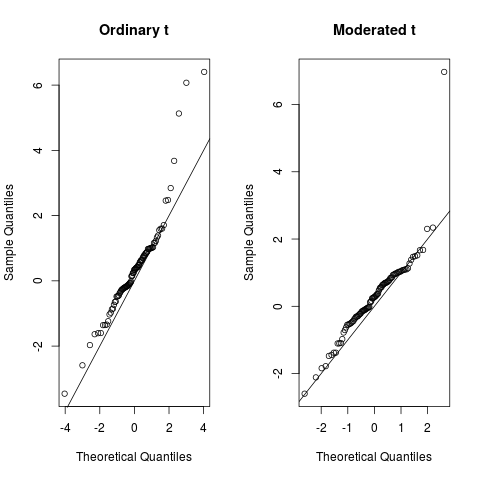
No counts, no variance: allowing for loss of degrees of freedom when assessing biological variability from RNA-seq data

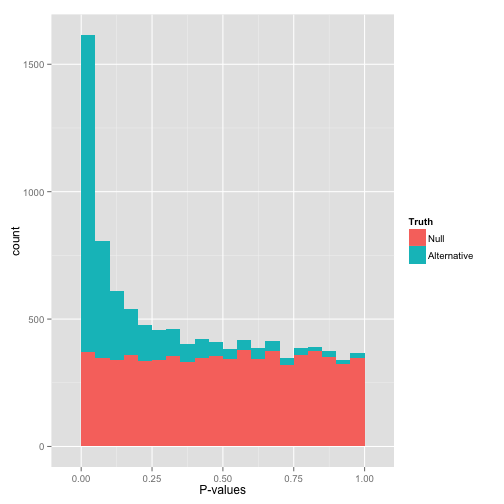
Histogram of p-values generated by conventional LIMMA and OSRR methods... | Download Scientific Diagram

Histogram of p-values of gene expression differences from duplicate... | Download Scientific Diagram

Histogram of p-values generated by conventional LIMMA and OSRR methods... | Download Scientific Diagram

Observation weights unlock bulk RNA-seq tools for zero inflation and single-cell applications. - Abstract - Europe PMC

Illustration how the global p-value is calculated. On the left ((a) and... | Download Scientific Diagram

Histogram of p-values generated by conventional LIMMA and OSRR methods... | Download Scientific Diagram

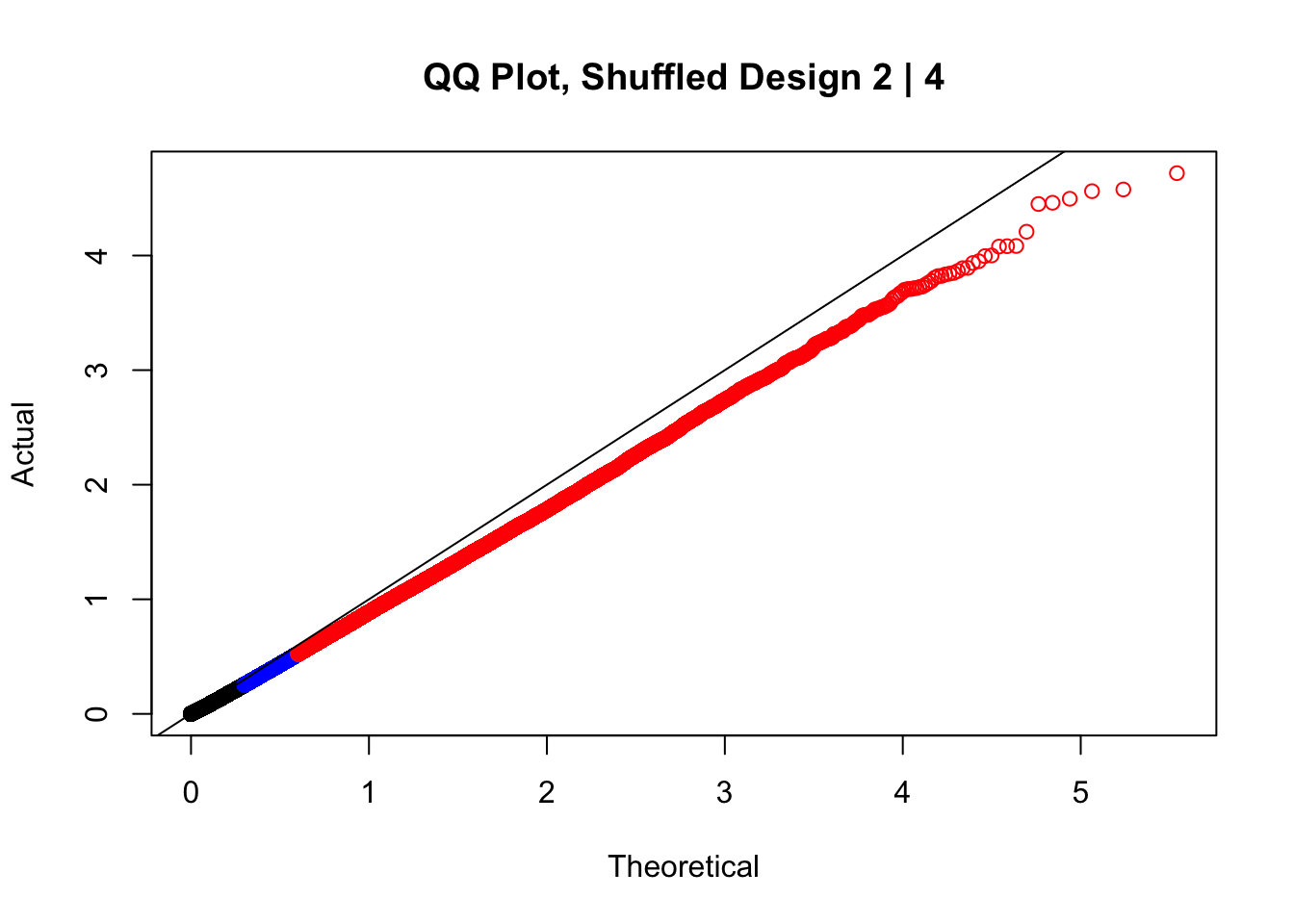
QQ plots of simulated null p-values for genes in TCGA HNSC study. (A)... | Download Scientific Diagram

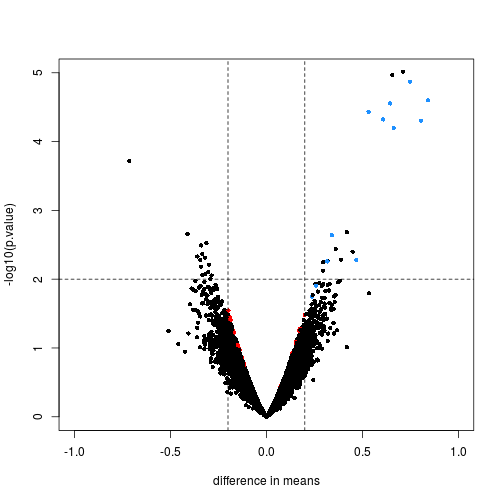
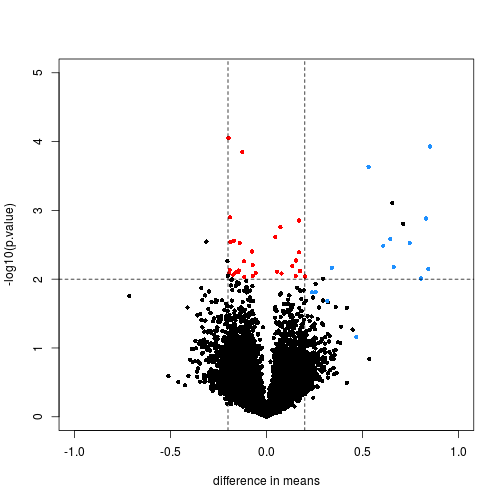
Controlling False Positive Rates in Methods for Differential Gene Expression Analysis using RNA-Seq Data | bioRxiv

dearseq: a variance component score test for RNA-Seq differential analysis that effectively controls the false discovery rate | bioRxiv